Welcome!
The Computational Neuroanatomy Group (CNG) is a laboratory dedicated to
the investigation of the structure, activity, and function of the nervous
system, from single cells to neuronal networks. Established in 1999, the
CNG (part of the Krasnow Institute for Advanced Study
at George Mason University) has been funded since
its inception by an R01 grant from NINDS and NIMH (and, from 2003, NSF),
under the Human Brain Project.
One of the main projects of CNG consists of the Generation and Description
of Dendritic Morphology. Dendrites have been qualitatively investigated
since the times of Golgi and Cajal. Only recently, however, has the use
of computer-interfaced microscopes allowed for the acquisition, storage,
and sharing of digital reconstructions of dendritic morphology. The opening
image of this document represents a detail of two Golgi-stained hippocampal
pyramidal cells traced and digitized in the CNG.
Here we provide a few examples of what has been achieved by the CNG
in the first four years of Human Brain Project support. Progress includes
the development of software for the quantitative analysis of dendritic
morphology, the implementation of computational models to simulate neuronal
structure, and the synthesis of anatomically accurate, large scale neuronal
assemblies in virtual reality.
 Analysis: The
availability of digitized neuronal reconstructions in principle allows
the extraction of any morphological measure from
single or multiple cells. We have developed L-Measure, the first freeware
software tool to quantitatively analyze dendritic morphology. After 2
years from the first release, L-Measure is used in more than a dozen laboratories
in the US and abroad. We recently employed L-Measure to carry out an extensive
statistical analysis of publicly available digitized CA3 and CA1 pyramidal
neurons. We found surprising differences, not only between the two classes,
but also between different reconstructing labs. For a
two-minute powerpoint show, click here (turn on the volume!).
Analysis: The
availability of digitized neuronal reconstructions in principle allows
the extraction of any morphological measure from
single or multiple cells. We have developed L-Measure, the first freeware
software tool to quantitatively analyze dendritic morphology. After 2
years from the first release, L-Measure is used in more than a dozen laboratories
in the US and abroad. We recently employed L-Measure to carry out an extensive
statistical analysis of publicly available digitized CA3 and CA1 pyramidal
neurons. We found surprising differences, not only between the two classes,
but also between different reconstructing labs. For a
two-minute powerpoint show, click here (turn on the volume!).
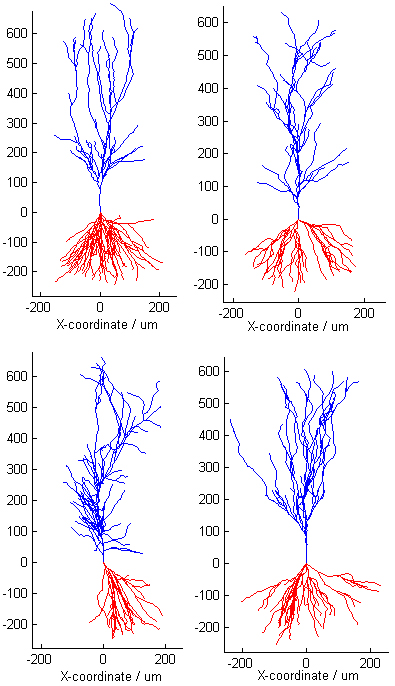
Synthesis: Based on biologically plausible "rules"
and biophysical determinants, we have designed stochastic models that
can generate realistic virtual neurons. Quantitative morphological analysis
indicates that virtual neurons are statistically compatible with the
real data that the model parameters are measured from. Here's a "turing
test": these 4 pyramidal neurons include 2 simulated and 2 real cells;
within each pair, one neuron is from the CA1 and one from the CA3 rat
hippocampus. Can you tell the real neurons from the virtual ones? (the
answer is at the end of this document...). Apical dendrites are blue, basal
are read. Click on the following animations to see the cells "grow" and
rotate in 3D. The file available formats are: animated gif, quicktime (mov),
mpeg (mp4), and flic (flc). Each format reproduces exactly the same animation
for each of the four cells.
Cell1.gif or cell1.mov or cell1.mp4 or cell1.flc
Cell2.gif or cell2.mov or cell2.mp4 or cell2.flc
Cell3.gif
or cell3.mov or cell3.mp4 or cell3.flc
Cell4.gif
or cell4.mov or cell4.mp4 or cell4.flc
Get the solution of the
Turing test (try to guess from the animations first!).
Networks: Virtual
neurons can be generated within an appropriate anatomical context if
a system level description of the surrounding tissue is included in the
model. As a first step towards a real-scale model of the hippocampus,
we have traced the  granule and molecular layers of the dentate
gyrus from microscopic MRI scans (click on the right MRI
image to see through the full stack), and arranged
2000 granule cells within the proper volume and with the correct orientation.
Finally, an axon reconstructed from the entorhinal cortex (part of the
perforant path, the main afferent to the dentate granule cells), has been
added in this virtual reality composition. To see the full animation, select
one of these movie formats:
granule and molecular layers of the dentate
gyrus from microscopic MRI scans (click on the right MRI
image to see through the full stack), and arranged
2000 granule cells within the proper volume and with the correct orientation.
Finally, an axon reconstructed from the entorhinal cortex (part of the
perforant path, the main afferent to the dentate granule cells), has been
added in this virtual reality composition. To see the full animation, select
one of these movie formats:
virtualDG1.mpg (mpeg1, medium resolution,
should run on all machines)
virtualDG2.mpg (mpeg2, higher resolution,
should run on most machines)
virtualDG3.avi (Microsoft Codec 9,
higher resolution, should run on Windows machines)
virtualDG4.avi (DVX Codec 5, highest
resolution, runs after free codec installation from divx.com)
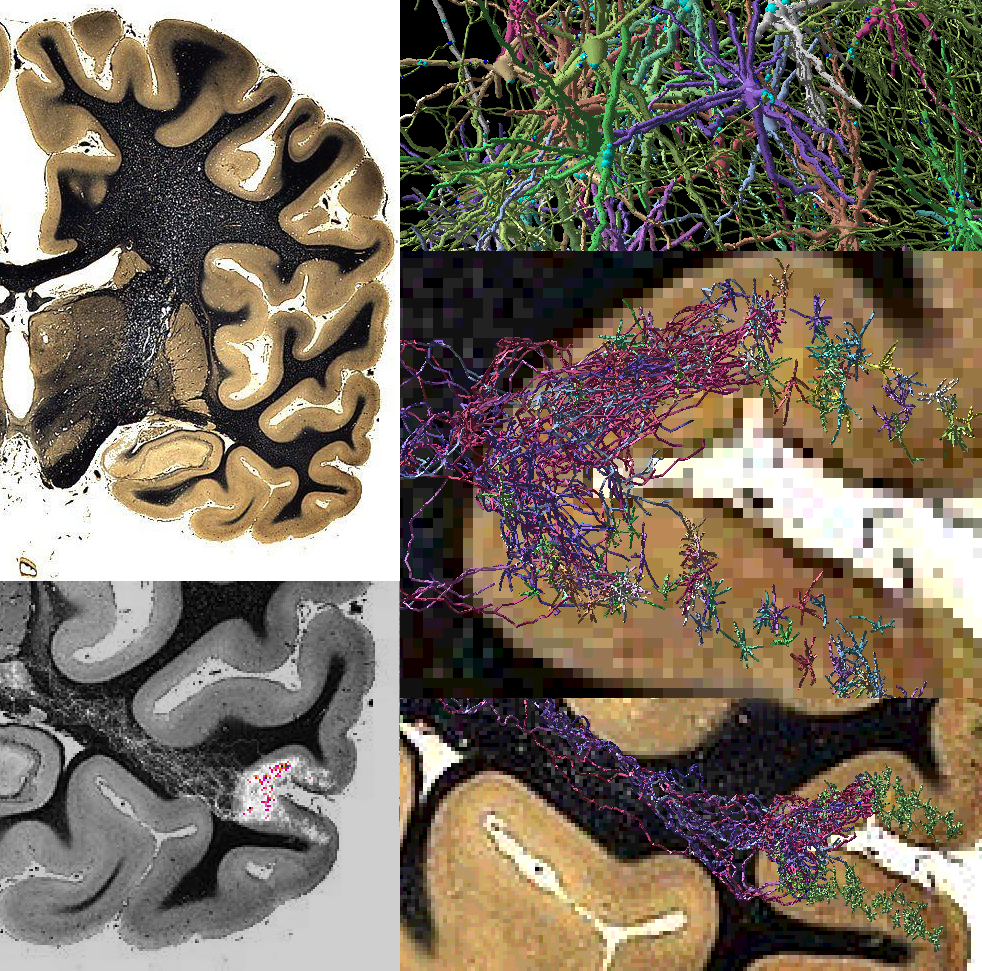
In order to simulate anatomically realistic neural
networks, axons must be grown as well as dendrites. We have developed
a navigation strategy for virtual axons in a voxel substrate. The panels
below (zooming in counter-clockwise from top left) are an example of simulated
axons stemming from virtual cortical cells and navigating towards the thalamus
through a substrate of voxels corresponding to a (real) stained section
of the human brain. Each virtual cell is assigned a different color.
Contact:
Giorgio Ascoli, ascoli@gmu.edu, Tel.
+1-703-993-4383
Links:
Computational Neuroanatomy
Group (follow links for more)
Human
Brain Project
Krasnow Institute for Advanced Study
George Mason University
Southampton Archive of Neuronal Morphology (and Cell Viewer)
Gulyas' Collection
(CA1 pyramidal cells and interneurons)
ImageJ
NeuroMorpho plugin
References:
BOOK: Ascoli G. (Ed.): Computational
Neuroanatomy - Principles and Methods (19 chapters, 468 pages, plus
CD-ROM). Humana Press, Totowa, NJ (2002).
Relevant Papers (1997-2003):
Ascoli G., Goldin R.: Coordinate systems for dendritic spines: a somatocentric
approach. Complexity 2(4):40-48 (1997).
Krichmar J., Ascoli G., Hunter L., Olds J.: A model of cerebellar saccadic
motor learning using qualitative reasoning. Lect. Notes Comp. Sci. 1240:134-145
(1997).
Vandersluis J., Cooke J., Ascoli G., Krichmar J., Michaels G., Montgomery
M., Symanzyk J., Vitucci B.: Exploratory statistical graphics for an initial
motion control experiment. Comp. Sci. Stat. 30:482-487 (1998).
Senft S., Ascoli G.: Reconstruction of brain networks by algorithmic
amplification of morphometry data. Lect. Notes Comp. Sci., 1606:25-33
(1999).
Ascoli G.: Progress and perspectives in computational neuroanatomy.
Anatom. Rec. 257(6):195-207 (1999).
Symanzik J, Ascoli G., Washington S., Krichmar J.: Visual data mining
of brain cells. Comp. Sci. Stat., 31:445-449 (1999).
Ascoli G., Krichmar J.: L-Neuron: a modeling tool for the efficient
generation and parsimonious description of dendritic morphology. Neurocomputing,
32-33:1003-1011 (2000).
Washington S., Ascoli G., Krichmar J.: A statistical analysis of dendritic
morphology’s effect on neuron electrophysiology of CA3 pyramidal cells.
Neurocomputing, 32-33:261-269 (2000).
Ascoli G.: The complex link between neuroanatomy and consciousness.
Complexity, 6(1):20-26 (2000).
Nasuto S., Krichmar J., Knape R., Ascoli G.: Relation between neuronal
morphology and electrophysiology in the kainate lesion model of Alzheimer's
Disease. Neurocomputing, 38-40:1477-1487 (2001).
Scorcioni R., Ascoli G.: Algorithmic extraction of morphological statistics
from electronic archives of neuroanatomy. Lect. Notes Comp. Sci., 2084:30-37
(2001).
Ascoli G., Krichmar J., Nasuto S., Senft S.: Generation, description,
and storage of dendritic morphology data. Phil. Trans. R. Soc. B, 356(1412):1131-45
(2001).
Ascoli G., Krichmar J., Scorcioni R., Nasuto S., Senft S.: Computer
generation and quantitative morphometric analysis of virtual neurons. Anat.
Embryol., 204:283-301 (2001).
Scorcioni R., Bouteiller J., Ascoli G.: A real-scale anatomical model
of the dentate gyrus based on single cell reconstructions and 3D rendering
of a brain atlas. Neurocomputing, 44-46:629-634 (2002).
Ascoli G.: Neuroanatomical algorithms for dendritic modeling. Network:
Comput. Neural Syst. 13:247-260 (2002).
Krichmar J., Nasuto S., Scorcioni R., Washington S., Ascoli G.: Effects
of dendritic morphology on CA3 pyramidal cell electrophysiology: a simulation
study. Brain Res., 941:11-28 (2002).
Lazarewicz M., Migliore M., Ascoli G.: A new bursting model of CA3
pyramidal cell physiology suggests multiple locations for spike initiation.
Biosystems, 67:129-37 (2002).
Samsonovich A., Ascoli G.: Statistical morphological analysis of hippocampal
principal neurons indicates selective repulsion of dendrites from their
own cell. J. Neurosci. Res. 71:173-87 (2003).
Gardner D., Toga A., Ascoli G., Beatty J., Brinkley J., Dale A., Fox
P., Gardner E., George J., Goddard N., Harris K., Herskovits E., Hines
M., Jacobs G., Jacobs R., Jones E., Kennedy D., Kimberg D., Mazziotta J.,
Miller P., Mori S., Mountain D., Reiss A., Rosen G., Rottenberg D., Shepherd
G., Smalheiser N., Smith K., Strachan T., Van Essen D., Williams R., Wong
S.: Sharing Data, Carefully. Neuroinformatics, 1:289-295 (2003).
Ascoli G.: Passive dendritic integration heavily affects spiking dynamics
of recurrent networks. Neural Networks, 16:657-663 (2003).
Scorcioni R., Lazarewicz M.T., Ascoli G.: Quantitative morphometry
of hippocampal pyramidal cells: differences between anatomical classes
and reconstructing laboratories. In Press, J. Comp. Neurol. (2004).
Relevant Book Chapters and Peer-reviewed Full-length Proceedings (1997-2003):
Krichmar J., Ascoli G., Olds J., Hunter L.: The qualitative reasoning
neuron: a new approach to modeling in computational neuroscience. In J.M.
Bower (Ed.): Computational Neuroscience: Trends in Research 1998, 609-614,
Plenum Press, New York, NY (1998).
Nasuto S., Krichmar J., Scorcioni R., Ascoli G.: Algorithmic statistical
analysis of electrophysiological data for the investigation of structure-activity
relationship in single neurons. InterJournal Complex Syst. R389:1-6 (2000).
Ascoli G.: Computing the brain and the computing brain. In G. Ascoli
(Ed.): Computational Neuroanatomy: Principles and Methods, 3-26, Humana
Press, Totowa, NJ (2002).
Donohue D., Scorcioni R., Ascoli G.: Generation and description of
neuronal morphology using L-Neuron: a case study. In G. Ascoli (Ed.):
Computational Neuroanatomy: Principles and Methods, 49-70, Humana Press,
Totowa, NJ (2002).
Lazarewicz M., Boer-Iwema S., Ascoli G.: Practical aspects in anatomically
accurate simulations of neuronal electrophysiology. In G. Ascoli (Ed.):
Computational Neuroanatomy: Principles and Methods, 127-148, Humana Press,
Totowa, NJ (2002).
Samsonovich A., Ascoli G.: Towards virtual brains. In G. Ascoli (Ed.):
Computational Neuroanatomy: Principles and Methods, 425-436, Humana Press,
Totowa, NJ (2002).
Turner DA, Cannon RC, Ascoli GA: Web-based neuronal archives:
neuronal morphometric and electrotonic analysis. In R. Kotter (Ed.): Neuroscience
Databases – A Practical Guide, 81-98, Elsevier, Amsterdam (2002).
Ascoli G., Samsonovich A.: Bayesian morphometry of hippocampal
cells suggests same-cell
somatodendritic repulsion. In Dietterich T.G., Becker S. Ghahramani
Z. (Eds.): Adv. Neural Proc. Syst. 14:133-139 (2002).
Ascoli G.: Electrotonic effects on spike response model dynamics. IEEE
Neural Networks, in press (2003).
Donohue D., Ascoli G.: Models of neuronal outgrowth. In Koslow S.H.
and Subramaniam S. (Eds.): Databasing the Brain: From Data to Knowledge,
Wiley, New York, NY. In Press (2004).
Credits:

This worked was supported by grant R01-39600 from NIMH, NINDS, and NSF
under the Human Brain Project of the Office of Neuroinformatics.
Current members of the Computational Neuroanatomy Group include:
Giorgio Ascoli, Principal Investigator
Xiaoshen Li, Postdoc
Alexei Samsonovich, Postdoc
Ruggero Scorcioni, Postdoc
Duncan Donohue, PhD Student
Deepak Ropyreddy, PhD Student
John Atkeson, MA Student
Sridevi Polavaram, MA Student
Kerry Brown, Pre-doc Student
David Velasquez, Pre-doc Student
Stephen Senft, Research Associate
In particular, material for this document has been contributed by
Alexei Samsonovich (Turing Test), Ruggero Scorcioni (Virtual Hippocampus),
and Steve Senft (Thalamocortical Projections).
This work would not be possible without the willingness of all active
neuroscientists to generously share their raw data and electronic tools
with the community. In particular, we are grateful to Drs. David Amaral,
German Barrionuevo, Jean-Marie Bouteiller, Gyuri Buzsaki, Robert Cannon,
Brenda Claiborne, Giampaolo D'Alessandro, Attila Gulyas, David Lester, Robert
Malenka, Michele Migliore, Nobu Tamamaki, and Dennis Turner.
Solution to the
Turing Test:
Cell1: Real CA3
Cell2: Virtual CA1
Cell4: Real CA1
Cell3: Virtual CA3

 Analysis: The
availability of digitized neuronal reconstructions in principle allows
the extraction of any morphological measure from
single or multiple cells. We have developed L-Measure, the first freeware
software tool to quantitatively analyze dendritic morphology. After 2
years from the first release, L-Measure is used in more than a dozen laboratories
in the US and abroad. We recently employed L-Measure to carry out an extensive
statistical analysis of publicly available digitized CA3 and CA1 pyramidal
neurons. We found surprising differences, not only between the two classes,
but also between different reconstructing labs. For a
two-minute powerpoint show, click here (turn on the volume!).
Analysis: The
availability of digitized neuronal reconstructions in principle allows
the extraction of any morphological measure from
single or multiple cells. We have developed L-Measure, the first freeware
software tool to quantitatively analyze dendritic morphology. After 2
years from the first release, L-Measure is used in more than a dozen laboratories
in the US and abroad. We recently employed L-Measure to carry out an extensive
statistical analysis of publicly available digitized CA3 and CA1 pyramidal
neurons. We found surprising differences, not only between the two classes,
but also between different reconstructing labs. For a
two-minute powerpoint show, click here (turn on the volume!).
 granule and molecular layers of the dentate
gyrus from microscopic MRI scans (click on the right MRI
image to see through the full stack), and arranged
2000 granule cells within the proper volume and with the correct orientation.
Finally, an axon reconstructed from the entorhinal cortex (part of the
perforant path, the main afferent to the dentate granule cells), has been
added in this virtual reality composition. To see the full animation, select
one of these movie formats:
granule and molecular layers of the dentate
gyrus from microscopic MRI scans (click on the right MRI
image to see through the full stack), and arranged
2000 granule cells within the proper volume and with the correct orientation.
Finally, an axon reconstructed from the entorhinal cortex (part of the
perforant path, the main afferent to the dentate granule cells), has been
added in this virtual reality composition. To see the full animation, select
one of these movie formats: